How to study the structure of complex biological samples?
High quality data and 3D envelopes can be obtained on small and large proteins with SEC-SAXS in the lab
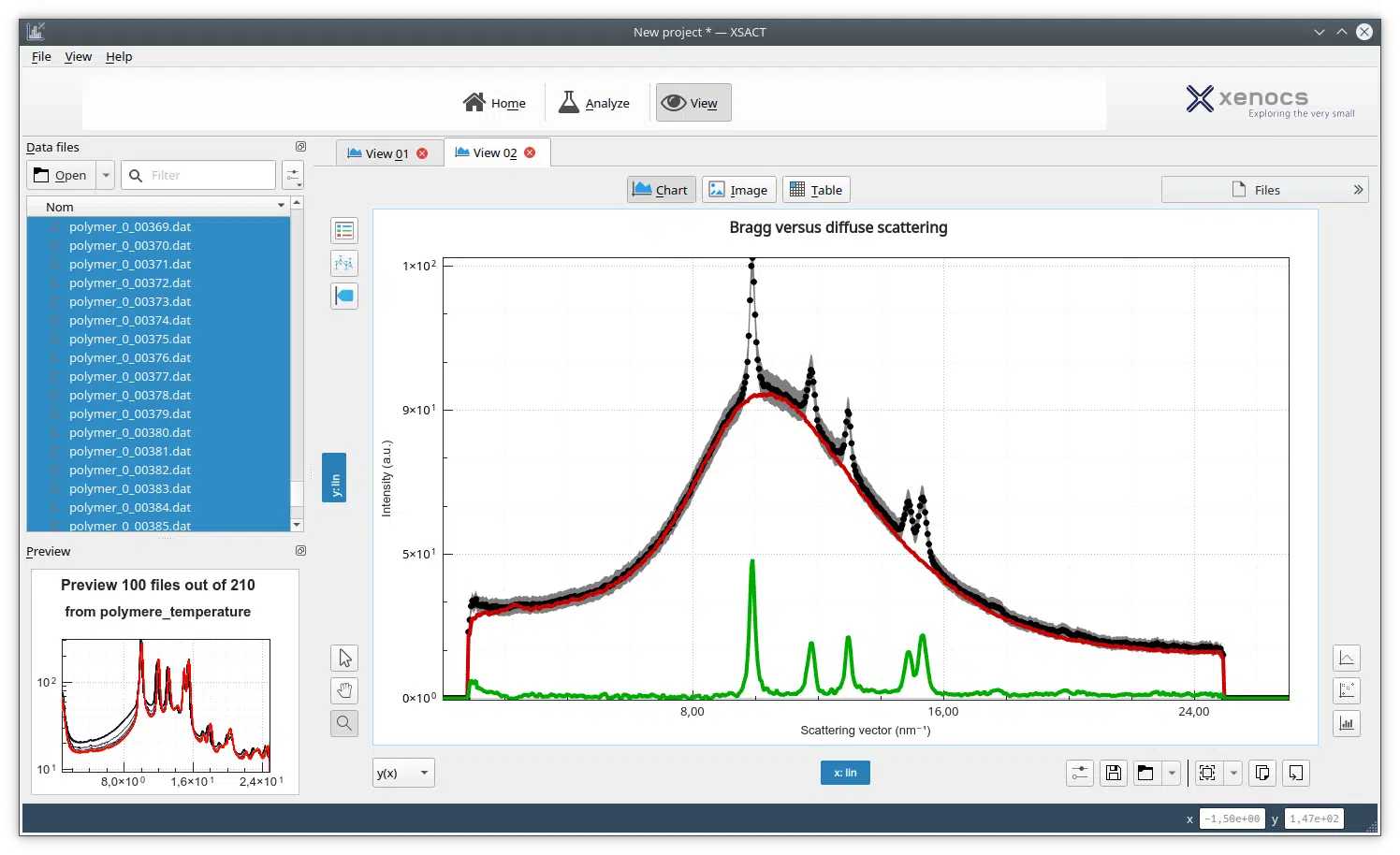
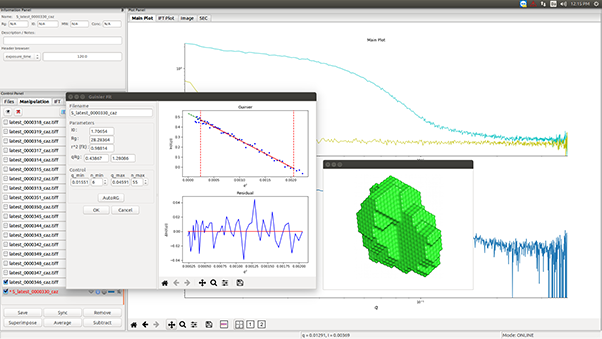
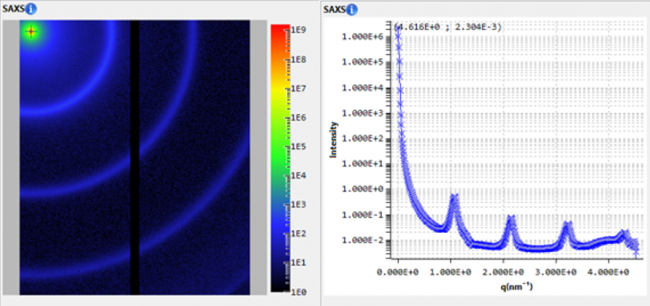
Small-angle X-ray scattering (SAXS) is commonly used to investigate the solution structure of biological macromolecules. Highly accurate results can be obtained with monodisperse solutions devoid of aggregates and impurities.
Many biological processes, however, involve structurally heterogeneous multi-domain proteins1 and heterogeneous protein complexes in dynamic equilibria2-6. While highly relevant, structural investigation of such samples is hampered by their transient or unstable nature. Size-Exclusion Chromatography (SEC) directly coupled with SAXS (SEC-SAXS) has emerged as a tool to mitigate these difficulties7-9.
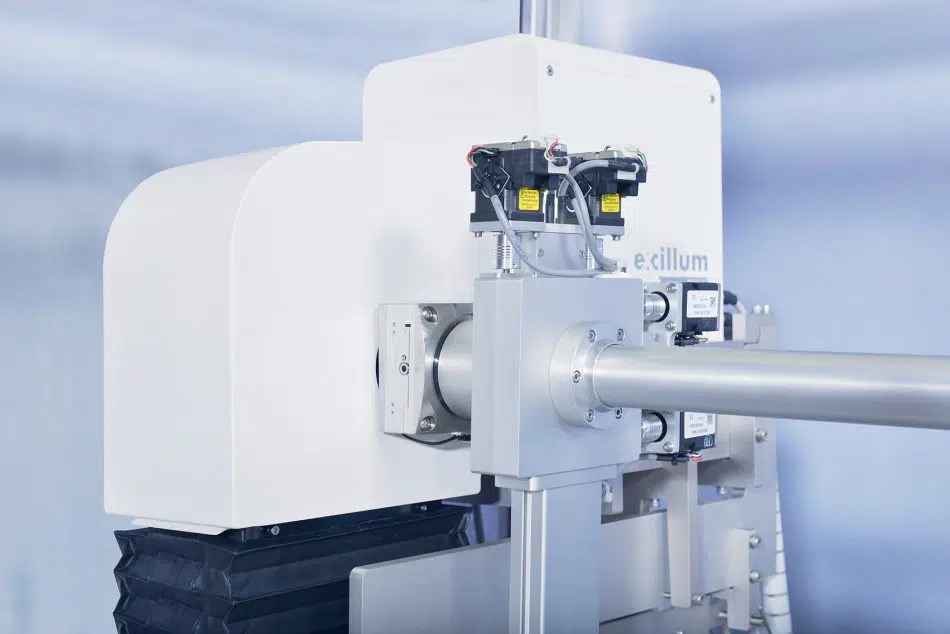
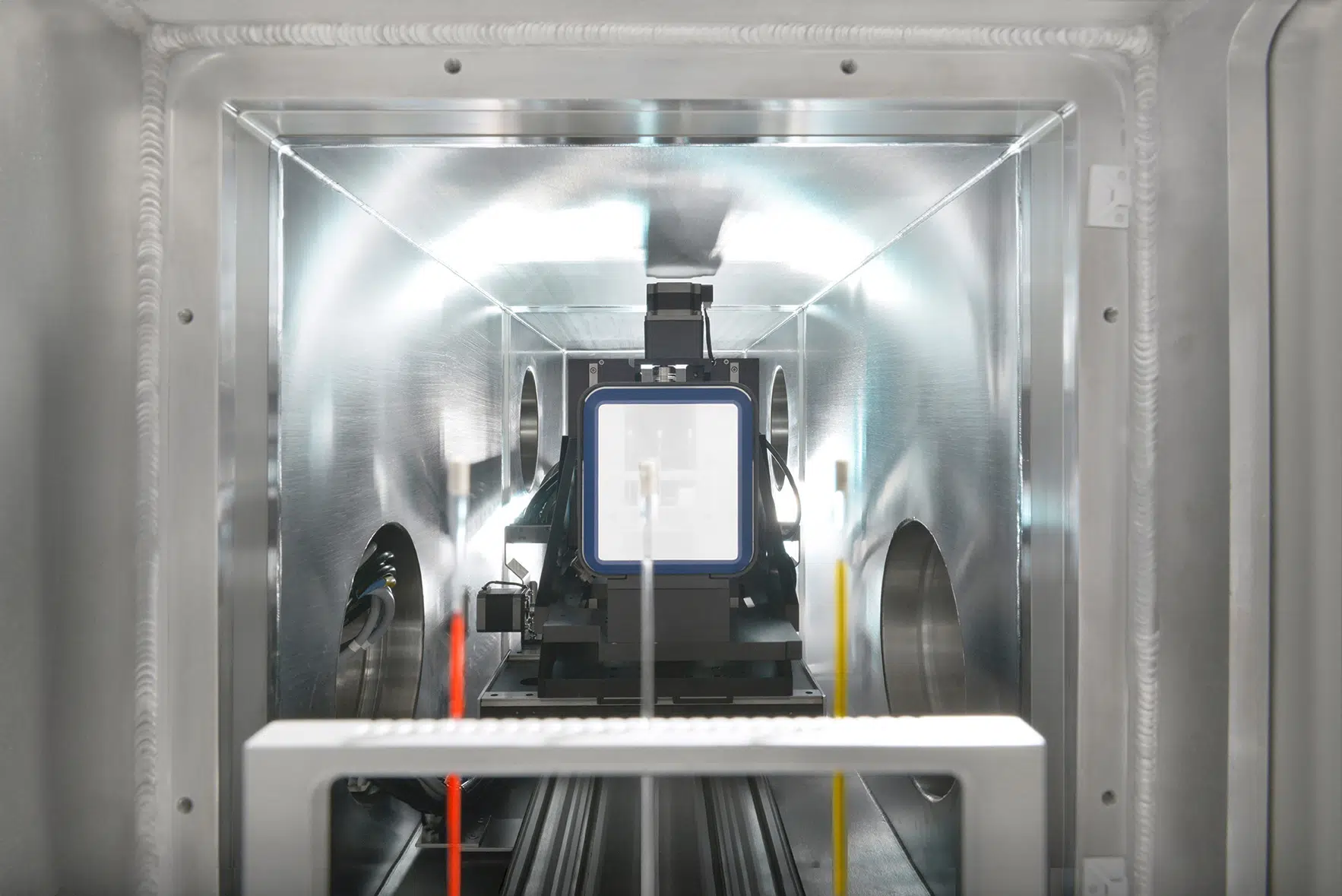
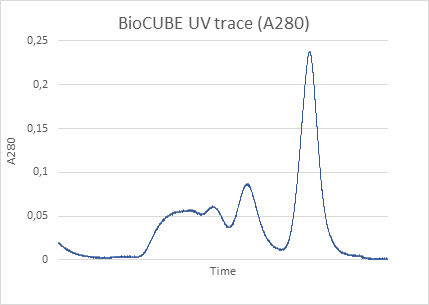
In the BioXolver SAXS instrument setup, samples eluting from a SEC column flows directly through a SAXS exposure capillary, allowing continuous data collection immediately following separation of individual components. SEC-SAXS furthermore ensures clean samples and removes potential work steps of sample fractionation and subsequent loading on the SAXS instrument for very stable samples, thus reducing sample consumption and agitation while facilitating and expediting the work-flow.
Want to read this application note?
In this application note you will discover:
- SEC-SAXS results on three different proteins, in different concentrations
- Structural investigation of minor solution components such as a small fraction of dimers in a mostly monomeric solution