Particle or Macromolecular Shape by SAXS
The three-dimensional shape of nanoparticles, proteins, micelles, and other nanoscale assemblies governs how they interact, assemble, transport, and function. Whether the structure is compact or elongated, globular or tubular, symmetric or anisotropic, its geometry affects reactivity, biological activity, colloidal stability, and mechanical or optical behavior.
Small-Angle X-ray Scattering (SAXS) provides a non-destructive and ensemble-averaged method to determine particle or macromolecular shape in solution or other native environments. SAXS analyzes the full scattering profile to reveal the distribution of distances within the particle. This allows reconstruction of its overall form without crystallization, labeling, or drying.
What is measured?
SAXS encodes structural information through the scattering intensity profile, which reflects the internal pairwise distances within the particle. Fourier transformation converts this information into the pair distance distribution function (PDDF), which represents all distances between points inside the particle.
From the PDDF, SAXS provides:
- Overall particle shape, distinguishing globular, rod-like, disk-like, or more complex geometries
- Characteristic dimensions, such as maximum particle dimension (Dmax), radii, or aspect ratios
- Internal symmetry and compactness, inferred from the PDDF profile shape
- Presence of extended or flexible domains, visible through long tails or broadened distributions
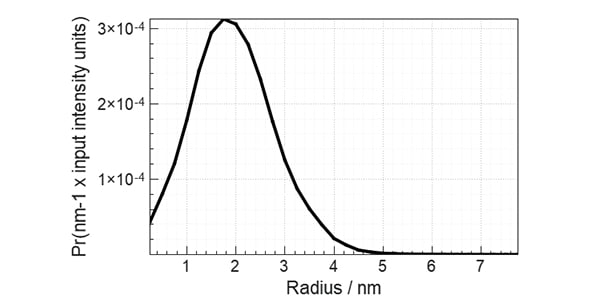
Figure 1. PDDF obtained from lysozyme solution SAXS data using XSACT Pro, showing a compact globular profile. PDDF is obtained through a Bayesian approach, with no information loss. The user input of auxiliary values such as the maximum distance within the particle (Dmax) of the smoothness parameter are not required.

Figure 2. Comparison between the PDDF of a spherical particle (40 nm in diameter) and a cylindrical particle (40 nm length and 4 nm diameter.), illustrating how shape information is encoded in real-space distance distributions.
XSACT Pro uses modern Bayesian approaches [1], to extract the PDDF with minimal user input and no loss of structural information, ensuring a robust reconstruction.
[1] S. Hansen, Bayesian estimation of hyperparameters for indirect Fourier transformation in small‐angle scattering, Journal of Applied Crystallography, 33.6 (2000). DOI: 10.1107/S0021889800012930.
Samples
SAXS shape determination applies to a wide range of nanoscale systems, particularly those that are monodisperse or moderately polydisperse.
Typical samples include:
- Protein solutions, including monomeric and oligomeric species
- Dilute nanoparticles in aqueous or organic media
- Surfactant assemblies and micelles, including block-copolymer micelles
- Lipid-based colloidal systems such as liposomes or vesicles
- Polymeric or inorganic nano-objects stable in suspension
Because researchers perform SAXS measurements in solution or in soft-matter environments, they preserve the native, hydrated structure of the particle.
Why use SAXS for Particle or Macromolecular Shape determination?
Ensemble-averaged statistical relevance
SAXS probes millions of particles within the illuminated volume, ensuring ensemble-averaged statistical relevance.
Non-destructive and label-free analysis
This preserves the sample’s native state.
Measurements under physiological or formulation conditions
Enabling shape determination in buffers, solvents, surfactant environments, or other realistic media.
Compatibility with flexible or partially disordered systems
Where high-resolution crystallography or microscopy may be limited.
Straightforward workflow
Allowing shape determination from a single scattering curve without the need for crystallization or staining.
These features make SAXS a powerful method for elucidating particle and macromolecular geometry in biostructural research, nanomaterial design, drug delivery systems, soft-matter physics, and colloidal science.