Molecular Weight by SAXS
Determining the molecular weight of macromolecules in solution is a key requirement in structural biology, biotechnology, pharmaceutical development, and soft-matter research. Molar mass provides essential information on oligomeric state, assembly, stability, and sample quality, helping researchers validate that molecules remain in their intended structural form.
Small-Angle X-ray Scattering (SAXS) offers a non-destructive and solution-based method to determine apparent molecular weight directly from the scattering intensity, without labeling, separation, or reference standards. Measurements are performed under conditions that preserve the native biochemical environment, revealing how molecules behave in realistic conditions.
What is measured?
SAXS detects X-rays scattered at small angles by electron density contrasts between solute molecules and solvent. In dilute solutions of globular macromolecules or proteins, the low-q region of the scattering curve contains information about overall particle size and forward scattering intensity, enabling direct estimation of the apparent molecular weight.
When intensities are expressed in absolute units, SAXS provides quantitative parameters, including:
- Apparent molecular weight, derived from the extrapolated forward intensity \( I(0) \)
- Mean molecular size or radius of gyration, determined through Guinier analysis [1]
- Sample concentration effects, incorporated naturally through absolute intensity normalization
Because SAXS collects scattering from entire molecular ensembles, the resulting molar mass estimate is statistically robust and representative of the actual population rather than a small subset of particles. The approach is valid for dilute, monodisperse samples where aggregation or interparticle interactions do not distort the low-q region.
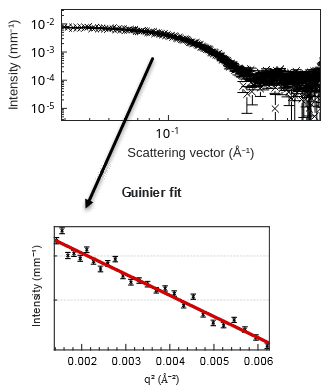
Figure 1. SAXS intensity curve of lysozyme in aqueous solution (top) and corresponding Guinier fit of the low-q region, processed with XSACT Pro software (bottom).
Modern laboratory SAXS instruments can reliably determine molecular weights in the 1 to ~300 kDa range.
[1] A. Guinier, G. Fournet, C.B. Walker, et al. Small‐angle scattering of X‐rays, (1956).
Samples
SAXS molecular weight determination applies to a wide range of globular macromolecules in solution, especially when the biological or physicochemical function depends on the oligomeric state or conformational integrity.
Typical sample types include:
- Globular proteins under native or buffer conditions
- Antibody fragments and protein complexes, including monomer–dimer equilibria
- Biological nano-objects such as lipoproteins or viral components
- Synthetic macromolecules designed to mimic protein-like scattering behavior
Because measurements occur directly in solution, SAXS preserves native folding, hydration, and interaction states, thus enabling reliable assessment of molecular weight in physiologically relevant environments.
Why use SAXS for Molecular Weight determination?
SAXS provides a robust and solution-compatible approach to determining the apparent molecular weight of macromolecules, complementing other structural and biophysical methods. Its main advantages include:
Non-destructive, native-state measurements
Enabling characterization in physiologically relevant buffers without altering molecular conformation.
Direct access to apparent molecular weight
Derived from absolute intensities without relying on reference proteins or external calibration standards.
Statistically representative analysis
Averaging scattering from the entire molecular ensemble and revealing aggregation or polydispersity when present.
Absolute intensity calibration
Enabling reliable determination of \( I(0) \) and apparent molecular weight.
Compatibility with in situ or time-resolved studies
Supporting investigations of structural stability, unfolding, or assembly dynamics in solution.
Additionally, SAXS can be coupled to separation methods like SEC (Size-Exclusion Chromatography) or AF4 (Asymmetric Flow Field-Flow Fractionation), allowing molecular weight determination on purified, monodisperse fractions. This combination minimizes aggregation effects and ensures that the low-q scattering reflects the true molecular species in solution.