“An in-house system provides the ability to respond in real time to the results, preparing new samples on the fly in response to what you are observing. This ability is often key to finding the right conditions for a particular sample.”
Associate Professor John Pascal
Department of Biochemistry and Molecular Medicine, Faculty of Medicine, Université de Montréal, Canada.
April 23, 2018
Interview with Associate Professor John Pascal, Department of Biochemistry and Molecular Medicine, Faculty of Medicine, Université de Montréal, Canada.
Could you please briefly introduce your research in structural biology?
J. Pascal : A broad research interest of my group is the cellular response to DNA damage. We study the proteins that detect and signal the presence of DNA damage, as well as the enzymes that repair the DNA damage. A rapid and efficient cellular response to DNA damage is critical to the integrity of the genome. Our research is aimed toward understanding how these proteins/enzymes work at the molecular level. We also have a strong interest in the family of proteins known as poly(ADP-ribose) polymerases, or PARPs. PARPs are involved in regulating virtually every aspect of human cell biology, and several PARP enzymes are targets for cancer treatment. Overall, we combine biochemical analysis, cell-based experiments, and structural biology techniques to advance our basic understanding of protein function, and we also work to leverage this structural information toward the development of new types of inhibitors that could potentially serve as therapeutics.
What are you using SAXS in your research for ?
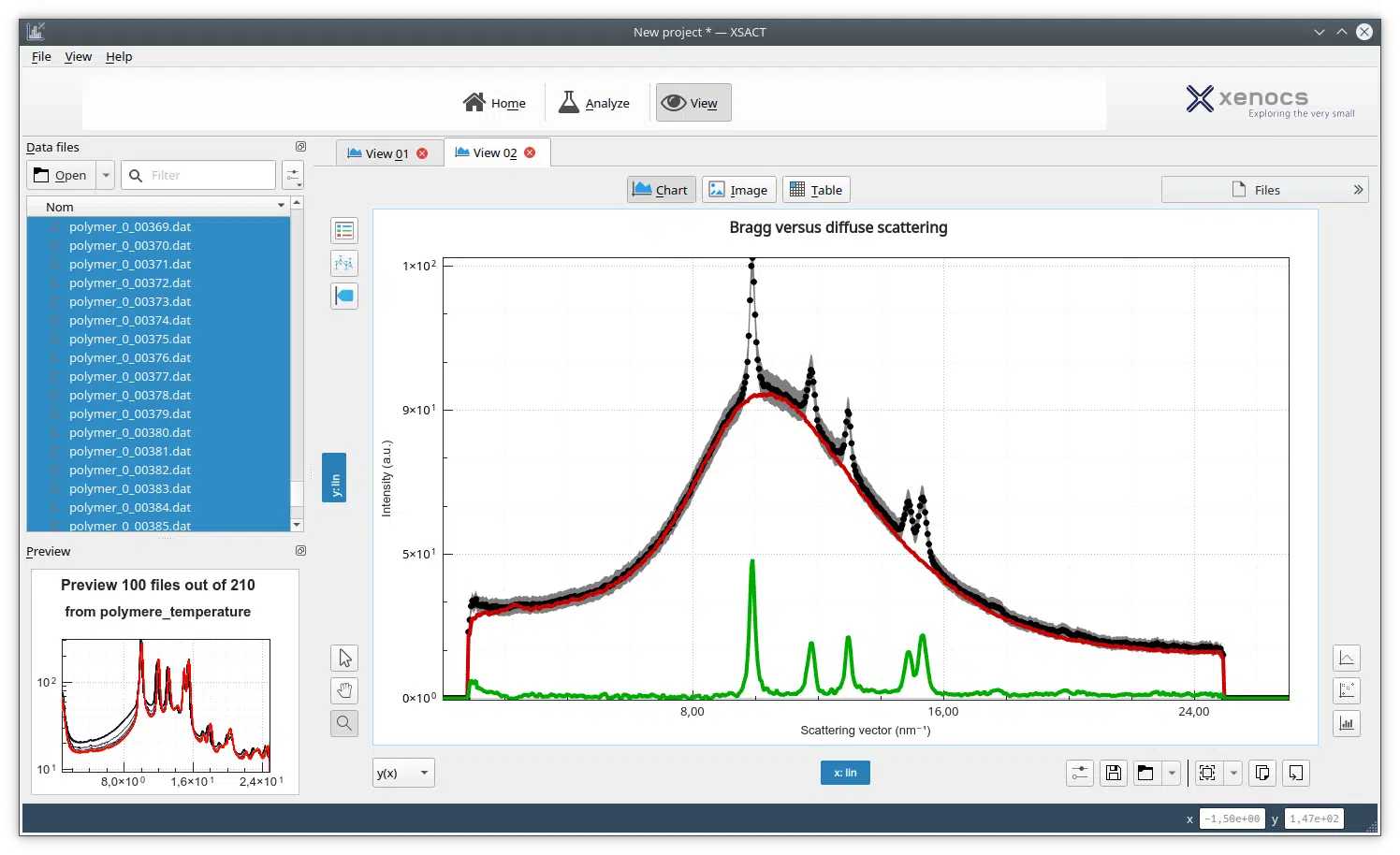
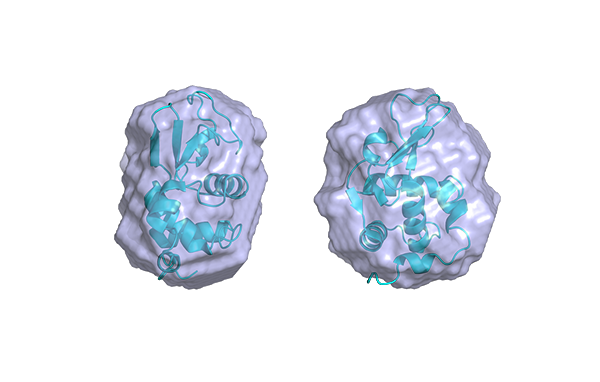
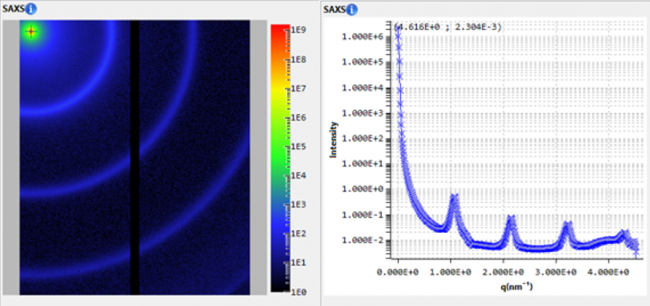
J. Pascal : We have used SAXS to address the overall shape and conformation of macromolecules and macromolecular complexes in solution. We are enthusiastic about SAXS! It has been particularly useful for the highly modular and flexible proteins that we have investigated. Whereas x-ray crystallography has often provided us with views of the more stable portions of a protein, SAXS analysis has often been able to capture information on full-length proteins and provide insights into the overall flexibility and relative positioning of the component parts/domains of the protein. We have also used SAXS to measure changes in protein conformation in response to interaction with small molecule inhibitors and protein binding partners.
What benefits does an in-house SAXS instrument have, compared with using a synchrotron facility?
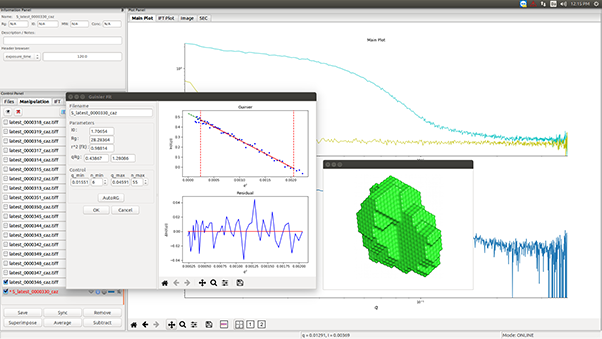
J. Pascal : “An in-house system provides the ability to respond in real time to the results, preparing new samples on the fly in response to what you are observing. This ability is often key to finding the right conditions for a particular sample.
There is also the question of how to ship the samples to the synchrotron, whether to send with a freeze then thaw cycle, or to just send as a fluid on ice. Not all samples behave well under these shipment conditions. The in-house source allows us to collect data when the sample is most “fresh”and ready-to-go. We have also used an in-house system to test for the best storage conditions so that we could then ship to synchrotrons more confidently.
Synchrotrons will always be an important and instrumental part of our work, but an in-house system helps us to properly prepare for optimal use of synchrotron facilities. We have had some proteins that suffer dramatically from synchrotron radiation damage, and an in-house source of lower intensity radiation has helped in these cases.”
Why did you choose to buy the BioXolver from SAXSLAB/Xenocs?
J. Pascal : “We liked that the capabilities of the system are quite adaptable to the needs of the experiment. We felt that we would never be limited in the types of experiments that we could perform.
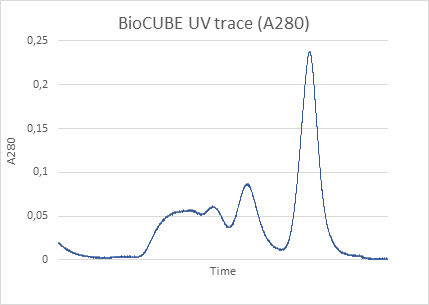
The machine is able to get through a substantial number of samples in a day’s time. The robotic sample loading and the visual feedback on sample positioning in the beamline are particularly nice. Precious and difficult to purify samples can be loaded in fairly small volumes that we can confidently keep in the beamline during the course of the experiment.”